INFOWEB 16 :
Aphyosemion revised and not split, with 2 new subgenera, Scheelsemion and Iconisemion
From Jean H. Huber
Private address: 7 Bd Flandrin, 75116 Paris, France
M.N.H.N., Ichthyology, PARIS, France.
Paris, February 3. 2014.
Dear Colleague, dear Aquarist!
As a researcher I have just published a few weeks ago a study with computerized systematics that, according to many immediate feedbacks and questions on systematics, often philosophical, has attracted a lot of interest.
Here is the abstract: The phylogeny of the African genus Aphyosemion (Cyprinodontiformes, Nothobranchiidae) is reappraised based on the single global molecular tree (Collier, 2007). A data matrix of 24 groups comprising all known subgenera and "orphan" species and 91 external characters (body and fins shape, color patterns) is produced that is congruent with the molecular tree. The subgenus Mesoaphyosemion is confirmed as polyphyletic, its definition is restricted and 2 new subgenera are described, Scheelsemion n. subgen. and Iconisemion n. subgen. with the previously assigned species to Mesoaphyosemion being redistributed to the "old" and the new subgenera. The allied genus Episemion is also confirmed as nested within the redefined genus Aphyosemion and is considered as its subgenus only, with its components possibly to be reconsidered once the area, rich of atypical ("orphan") species is fully sampled.
Why are the generic changes not automatically followed by Killi-Data like in the past ?
This is the most frequently asked question to Killi-Data (see the newsletters 12 and 13), and not only for the Rivulus and the Simpsonichthys cases. Here with Aphyosemion the question is raised again.
Has Killi-Data become over-conservative or rigid or even personal-minded or trying to impose a vision to everybody else, notably baring in mind that precisely Killi-Data had started in 2000 by pioneeringly stating that changes at the generic or species level will always be automatically followed if evidence based… therefore why not follow the split of Rivulus and Simpsonichthys and Aphyosemion ?
The answer is simple and just a fact of life.
In the past that pioneering -and objective- principle was thought necessary because conflicts between splitters and lumpers were culminating and they were based on opinions only (remember the Roloffia case, no matter of the I.C.Z.N. further decision, that is restricted to nomenclatural issues). Then knowledge and evidence-based publications have increased sharply and a steady and growing tendency to split everything has influenced the publications everywhere and also among killifish (notably with the emergence of the Brasilian researcher Wilson Costa who has described about 230 killifish names to-date in about 25 years).
With these many many names we have a mirror of human life for any system (here a sound principle, to produce and respect evidence based data) that becomes emptied of its sense (perverted, may say some ironists) over time and, for example, Wilson Costa has (no critics, nothing personal against him here, just his strategy as a researcher) changed the picture of the old single genus Cynolebias (sensu Parenti, 1981) into at least 12 genera that were up-levelled from previous subgenera or created as atypical genera (Cynolebias, Austrolebias, Simpsonichthys, Hypsolebias, Nematolebias, Ophthalmolebias, Spectrolebias, Xenurolebias, Plesiolebias, Stenolebias, Maratecoara, Papiliolebias) some of them being further divided in several subgenera (e.g., Austrolebias in 7 subgenera), pending the next step of uplevelling these present subgenera to genera again, and similarly for Cynopoecilus and for Rivulus, while the publications to sustain these splits appeared more as preliminary steps into improving progressively a poor status of knowledge (in other words, proposed diagnostic characters appeared not solid enough or roughly analysed or too quickly translated into generic names).
That is the main reason why Killi-Data has not followed the split of Rivulus and Simpsonichthys with new genera and may even reverse the picture in other also over-splitted groups, after new publications (and in parallel several scientific websites on fishes -not only on killifish- have just done the same).
Nothing justifies that extreme and repetitive splitting strategy in the concerned groups (except a subjective philosophy that is respectable but not due to everybody) while in other groups, other researchers with also modern technology and evidence-based techniques have moved the other way by lumping subgenera into synonyms or by keeping subgenera at the same level (e.g., for Adinia and Fundulus, in 2013 or for Rivulus and its allied, in 2012, op. cit., or by maintaining untouched Aphyosemion as a speciose genus). On the other hand, that position by Killi-Data may bear some weaknesses : it refrains in some way the acknowledgements of new names by the scientific community and it may be seen as unbalanced between new researchers that still must have the right to describe new names at the generic level and old researchers who did it in the past (and had the single advantage to have been born earlier !)
Indeed, the case of the genus Aphyosemion is emblematic because several subgenera had been described in the nineteen seventies and some authors have been tempted to up-level them as genera (e.g., Chromaphyosemion, Diapteron, Raddaella) and they even proposed it but they were poorly followed because nobody could handle the case of the subgenus Mesoaphyosemion that molecular data had clearly shown as heterogeneous (poly- or paraphyletic) and everybody knew that the morphology of all Aphyosemion was extremely stable (and ironically the single lineage that could be told apart, Raddaella fell close to Fundulopanchax by morphology, but not at all by DNA !)… in other words it was not reasonable (and would have been harshly criticised) to split some subgenera into genera and leave Mesoaphyosemion untouched.
As properly stated by Collier (2007) : a possible splitting of the genus in several genera is not advisable if the concept of genus proposed by him is followed ("If genera are to reflect phylogenetic history, two conditions must be met. First, all members of a genus should be descended from a single common ancestor. Second, all descendants of that common ancestor should be members of the same genus") and besides differences beween subgeneric lineages are not big too (as an additional argument) and besides, even more (cf. Pauciradius), they are not big between some subgenera of Aphyosemion and some subgenera of Fundulopanchax !
Why is it necessary to mimic molecular data to build in the genus Aphyosemion ?
Collier's molecular results (2007) may be summarized by the following items :
- all studied populations/species (over 100) described within the genus Aphyosemion are phylogenetically related in the tree (hence the genus is monophyletic);
- the studied component of Episemion is not placed outside the genus Aphyosemion in the molecular tree, but inside, and should not be considered as a distinct genus but since its studied component is placed closest to atypical species with weakly solid branches a formal synonymisation of Episemion is not formally requested;
- there are, within the tree, several species named by Collier as "orphans" (hera, callipteron, hofmanni) that are not positioned firmly in the tree (i.e., they are included in subbranches with low bootstrap values, in-between the 2 main solid branches);
- there are, within the tree, several species that were initially described as atypical based on morphological characters and that are solidly placed in the tree within unexpected species groups (e.g., herzogi, tirbaki, labarrei, hanneloreae, joergenscheeli);
- the sharply diagnosed subgenera of Aphyosemion – namely Chromaphyosemion, Kathetys, Raddaella and Diapteron – are confirmed as corresponding to well homogeneous phylogenetic and individualized lineages;
- the not sharphy diagnosed subgenus Mesoaphyosemion, comprising species with average morpho-meristics is not monophyletic and needs further work.
As a direct consequence, it appeared to the author (and previously probably to Collier who did not change the systematics of the genus apart from suggesting the inclusion of Episemion into it after his molecular results) that it was hopeless to produce a purely morphological tree from mainly measurable data (proportions and meristics) like it had been achieved for the Rivulins (Huber, 1999, 2012, Costa, 2006, 2011) or for the lampeyes (Huber, 1999, 2011). It was then decided to try to mimic the results of the molecular tree (Collier, 2007, and also Agnèse, Sonnenberg, Zentz, on more focused subgroups of Aphyosemion) by using morphological data and notably details of the live color pattern and to uniquely declare that option with a formal statement (although it cannot be said for sure that no researcher has not used molecular trees in the past in order to decide on which morphological data to use, none has formally declared it as such, to-date). In addition, after Collier's work (2007), our knowledge was still left with a paraphyletic subgenus, Mesoaphyosemion, and with old diagnoses for all other named subgenera. It was then thought useful and fully in agreement with ICZN which requires formal diagnosis attached to generic levels, to try to mimic the molecular tree to develop new diagnoses for old and possibly new subgenera : that was achieved after the analysis of 91 characters (see the following morphological tree) but reviewers asked not to go further (notably on bootstrap values) because of the "a priori" conception of the tree, unlike previous works on Rivulins and lampeyes, also as computerized systematics. Not only the obtained tree was mimicing the molecular tree but it confirmed the position of the atypical species (point 4, above) and even helped to clarify the position of Collier's "orphan" species [note : several aquarists ask after comparing the present tree with Collier's tree about differences, e.g., about the relative position of Episemion subbranch not bordering that of Diapteron… this is only apparent because the computer decides where to place visually a subbranch relatively to its next subbranch, above or below, but the 2 subbranches can easily be reversed ("rolled") without changing the tree (see also the last paragraph on future studies)].
As a consequence, the new systematic definition of the genus Aphyosemion and its subgenera is fully congruent with present molecular evidence. It comprises 9 subgenera (for more than a hundred species) and it is fully homogeneous and stable.
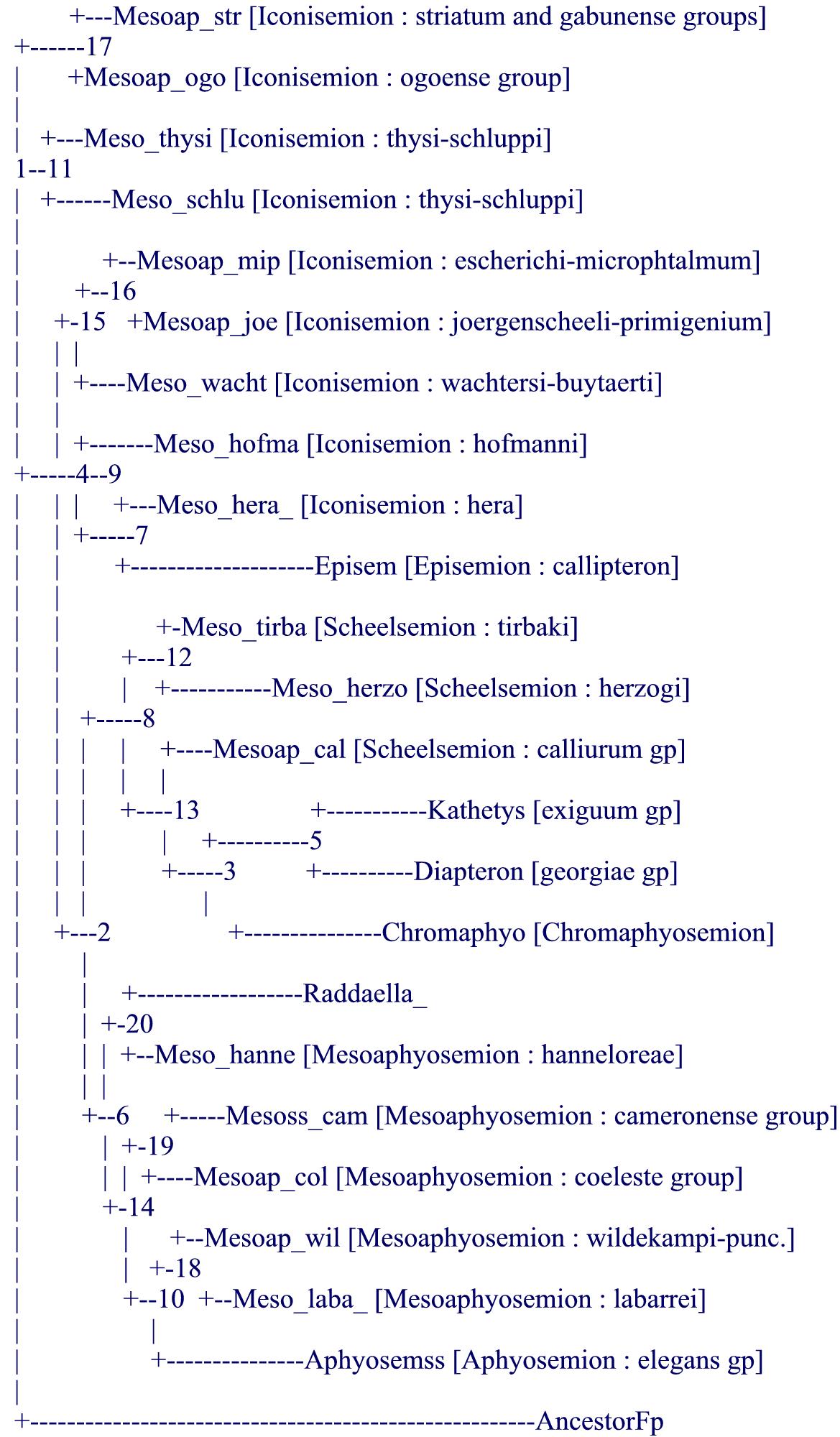 .
.
What are the impacts of the reshuffling of the subgenera of Aphyosemion ?
Here is a table that summerizes the situation after the changes
notes : in the article the status of wuendschi as a subspecies of hanneloreae is questioned but not resolved (possibly closer to hofmanni ?) because the author has never seen that fish ; there is a minor copy-paste error in the publication : labarrei is listed in Iconisemion while it is clearly among Mesoaphosemion in its new restricted sense in the computerized tree, and also in the molecular tree (and the error has been corrected herein).
 LIST OF CURRENT NAMES WITH ASSIGNED SUBGENERA
LIST OF CURRENT NAMES WITH ASSIGNED SUBGENERA
| FULL NAME | ABBREVIATED NAME | CURRENT STATUS | ALTERNATIVE STATUS |
|---|---|---|---|
| Aphyosemion (Diapteron) abacinum | A. (D.) abacinum | valid sp. | |
| Aphyosemion (Scheelsemion) ahli | A. (Sch.) [Sch.cal] ahli | valid sp. | |
| Aphyosemion (Chromaphyosemion) alpha | A. (Chrom.) alpha | valid sp. | |
| Aphyosemion (Mesoaphyosemion) amoenum | A. (Mes.) [Mes.cam] amoenum | valid sp. | # subsp. amoenum amoenum |
| Aphyosemion (Mesoaphyosemion) aureum | A. (Mes.) [Mes.col] aureum | valid sp. | |
| Aphyosemion (Scheelsemion) australe | A. (Sch.) [Sch.cal] australe | valid sp. | |
| Aphyosemion (Kathetys) bamilekorum | A. (Kath.) bamilekorum | valid sp. | |
| Aphyosemion (Raddaella) batesii | A. (Rad.) batesii | valid sp. | |
| Aphyosemion beauforti | A. (Rad.) beauforti | = batesii | # = cameronense |
| Aphyosemion bellicauda | A. (Mes.) [Mes.cam] bellicauda | = cameronense | |
| Aphyosemion (Chromaphyosemion) bitaeniatum | A. (Chrom.) bitaeniatum | valid sp. | |
| Aphyosemion (Chromaphyosemion) bivittatum | A. (Chrom.) bivittatum | valid sp. | |
| Aphyosemion bochtleri | A. (Sch.) [Sch.hez] bochtleri | = herzogi | # subsp. herzogi |
| Aphyosemion (Iconisemion) boehmi | A. (Icon.) [Icon.str] boehmi | valid sp. | # subsp. gabunense |
| Aphyosemion (Kathetys) bualanum | A. (Kath.) bualanum | valid sp. | # subgen. Mesoaphyosemion |
| Aphyosemion (Iconisemion) buytaerti | A. (Icon.) [Icon.wac] buytaerti | valid sp. | |
| Aphyosemion caeruleum Meinken [non Boulenger] | A. (Sch.) [Sch.cal] caeruleum Meinken [non Boulenger] | nom. preoc. = ahli | |
| Aphyosemion (Episemion) callipteron | A. (Epis.) callipteron | valid sp. | |
| Aphyosemion (Scheelsemion) calliurum | A. (Sch.) [Sch.cal] calliurum | valid sp. | |
| Aphyosemion (Mesoaphyosemion) cameronense | A. (Mes.) [Mes.cam] cameronense | valid sp. | # subsp. cameronense cameronense |
| Aphyosemion (Scheelsemion) campomaanense | A. (Sch.) [Sch.cal] campomaanense | valid sp. | |
| Aphyosemion carnapi | A. (Kath.) carnapi | = exiguum | |
| Aphyosemion (Aphyosemion) castaneum | A. (A.) castaneum | valid sp. | # = christyi |
| Aphyosemion (Iconisemion) caudofasciatum | A. (Icon.) [Icon.ogo] caudofasciatum | valid sp. | # subsp. ogoense |
| Aphyosemion (Scheelsemion) celiae celiae | A. (Sch.) [Sch.cal] celiae | nomin. subsp. celiae celiae | |
| Aphyosemion (Aphyosemion) chauchei | A. (A.) chauchei | valid sp. | |
| Aphyosemion (Aphyosemion) christyi | A. (A.) christyi | valid sp. | |
| Aphyosemion (Mesoaphyosemion) citrineipinnis | A. (Mes.) [Mes.col] citrineipinnis | valid sp. | |
| Aphyosemion (Mesoaphyosemion) coeleste | A. (Mes.) [Mes.col] coeleste | valid sp. | |
| Aphyosemion coeruleum Meinken [non Boulenger] | A. (Chrom.) coeruleum Meinken [non Boulenger] | nom. preoc. = hollyi | |
| Aphyosemion (Aphyosemion) cognatum | A. (A.) cognatum | valid sp. | |
| Aphyosemion (Aphyosemion) congicum | A. (A.) congicum | valid sp. | # nom. dubium |
| Aphyosemion (Diapteron) cyanostictum | A. (D.) cyanostictum | valid sp. | |
| Aphyosemion (Kathetys) dargei | A. (Kath.) dargei | valid sp. | |
| Aphyosemion (Aphyosemion) decorsei | A. (A.) decorsei | valid sp. | |
| Aphyosemion (Chromaphyosemion) ecucuense | A. (Chrom.) ecucuense | valid sp. | # = malumbresi |
| Aphyosemion (Scheelsemion) edeanum | A. (Sch.) [Sch.cal] edeanum | valid sp. | |
| Aphyosemion elberti | A. (Kath.) elberti | = bualanum | # valid sp. |
| Aphyosemion (Aphyosemion) elegans | A. (A.) elegans | valid sp. | |
| Aphyosemion (Chromaphyosemion) erythron | A. (Chrom.) erythron | valid sp. | |
| Aphyosemion (Iconisemion) escherichi | A. (Icon.) [Icon.str] escherichi | valid sp. | # = striatum |
| Aphyosemion (Mesoaphyosemion) etsamense | A. (Mes.) [Mes.cam] etsamense | valid sp. | |
| Aphyosemion (Iconisemion) exigoideum | A. (Icon.) [Icon.str] exigoideum | valid sp. | |
| Aphyosemion (Kathetys) exiguum | A. (Kath.) exiguum | valid sp. | |
| Aphyosemion (Aphyosemion) ferranti | A. (A.) ferranti | valid sp. | |
| Aphyosemion (Scheelsemion) pascheni festivum | A. (Sch.) [Sch.pas] festivum | subsp. pascheni | # valid sp. |
| Aphyosemion flavum | A. (Sch.) [Sch.cal] flavum | = calliurum | |
| Aphyosemion (Scheelsemion) franzwerneri | A. (Sch.) [Sch.fra] franzwerneri | valid sp. | |
| Aphyosemion (Diapteron) fulgens | A. (D.) fulgens | valid sp. | |
| Aphyosemion (Iconisemion) gabunense | A. (Icon.) [Icon.str] gabunense | valid sp. | # subsp. gabunense |
| Aphyosemion (Diapteron) georgiae | A. (D.) georgiae | valid sp. | |
| Aphyosemion (Iconisemion) grelli | A. (Icon.) [Icon.hof] grelli | valid sp. | |
| Aphyosemion gustavi | A. (Rad.) gustavi | = batesii | |
| Aphyosemion (Mesoaphyosemion) haasi | A. (Mes.) [Mes.cam] haasi | valid sp. | # subsp. cameronense |
| Aphyosemion (Mesoaphyosemion) halleri | A. (Mes.) [Mes.cam] halleri | valid sp. | # subsp. cameronense |
| Aphyosemion (Mesoaphyosemion) hanneloreae hanneloreae | A. (Mes.) [Mes.han] hanneloreae | nomin. subsp. hanneloreae hanneloreae | |
| Aphyosemion (Scheelsemion) heinemanni | A. (Sch.) [Sch.cal] heinemanni | valid sp. | |
| Aphyosemion (Iconisemion) hera | A. (Icon.) [Icon.her] hera | valid sp. | |
| Aphyosemion (Scheelsemion) herzogi | A. (Sch.) [Sch.hez] herzogi | valid sp. | # subsp. herzogi herzogi |
| Aphyosemion (Iconisemion) hofmanni | A. (Icon.) [Icon.hof] hofmanni | valid sp. | |
| Aphyosemion hollyi | A. (Chrom.) hollyi | = bivittatum | |
| Aphyosemion jacobi | A. (Kath.) jacobi | = exiguum | |
| Aphyosemion jaundense | A. (Kath.) jaundense | = exiguum | |
| Aphyosemion (Iconisemion) joergenscheeli | A. (Icon.) [Icon.str] joergenscheeli | valid sp. | |
| Aphyosemion (Kathetys) kekemense | A. (Kath.) kekemense | valid sp. | # subsp. bualanum # = elberti |
| Aphyosemion (Chromaphyosemion) kouamense | A. (Chrom.) kouamense | valid sp. | |
| Aphyosemion (Chromaphyosemion) koungueense | A. (Chrom.) koungueense | valid sp. | |
| Aphyosemion (Episemion) krystallinoron | A. (Epis.) krystallinoron | valid sp. | |
| Aphyosemion kunzi | A. (Rad.) kunzi | = splendidum | # = batesii # valid sp. |
| Aphyosemion (Mesoaphyosemion) labarrei | A. (Mes.) [Mes.wil] labarrei | valid sp. | |
| Aphyosemion (Aphyosemion) lamberti | A. (A.) lamberti | valid sp. | |
| Aphyosemion (Aphyosemion) lefiniense | A. (A.) lefiniense | valid sp. | # subsp. schioetzi |
| Aphyosemion (Scheelsemion) lividum | A. (Sch.) [Sch.cal] lividum | valid sp. | |
| Aphyosemion loboanum | A. (Kath.) loboanum | = exiguum | |
| Aphyosemion (Chromaphyosemion) loennbergii | A. (Chrom.) loennbergii | valid sp. | |
| Aphyosemion loloense | A. (Kath.) loloense | = exiguum | |
| Aphyosemion (Iconisemion) louessense | A. (Icon.) [Icon.ogo] louessense | valid sp. | |
| Aphyosemion (Chromaphyosemion) lugens | A. (Chrom.) lugens | valid sp. | |
| Aphyosemion (Aphyosemion) lujae | A. (A.) lujae | valid sp. | |
| Aphyosemion (Mesoaphyosemion) maculatum | A. (Mes.) [Mes.cam] maculatum | valid sp. | |
| Aphyosemion (Chromaphyosemion) malumbresi | A. (Chrom.) malumbresi | valid sp. | |
| Aphyosemion margaretae | A. (A.) margaretae | = christyi | # valid sp. |
| Aphyosemion (Iconisemion) marginatum | A. (Icon.) [Icon.str] marginatum | valid sp. | # subsp. gabunense |
| Aphyosemion (Chromaphyosemion) melanogaster | A. (Chrom.) melanogaster | valid sp. | |
| Aphyosemion melanopteron | A. (A.) melanopteron | = congicum | # valid sp. |
| Aphyosemion (Chromaphyosemion) melinoeides | A. (Chrom.) melinoeides | valid sp. | |
| Aphyosemion microphtalmum | A. (Icon.) [Icon.str] microphtalmum | = escherichi | # valid sp. |
| Aphyosemion microstomum | A. (Mes.) [Mes.cam] microstomum | = cameronense | |
| Aphyosemion mikeae | A. (Icon.) [Icon.wac] mikeae | = wachtersi | # subsp. wachtersi |
| Aphyosemion (Mesoaphyosemion) mimbon | A. (Mes.) [Mes.cam] mimbon | valid sp. | |
| Aphyosemion multicolor | A. (Chrom.) multicolor | = bitaeniatum | # valid sp. |
| Aphyosemion (Aphyosemion) musafirii | A. (A.) musafirii | valid sp. | |
| Aphyosemion nigri | A. (Chrom.) nigri | = bitaeniatum | |
| Aphyosemion normani | A. (Kath.) normani | = exiguum | |
| Aphyosemion (Mesoaphyosemion) obscurum | A. (Mes.) [Mes.cam] obscurum | valid sp. | # subsp. cameronense |
| Aphyosemion (Mesoaphyosemion) ocellatum | A. (Mes.) [Mes.col] ocellatum | valid sp. | |
| Aphyosemion (Iconisemion) ogoense | A. (Icon.) [Icon.ogo] ogoense | valid sp. | # subsp. ogoense ogoense |
| Aphyosemion (Chromaphyosemion) omega | A. (Chrom.) omega | valid sp. | |
| Aphyosemion (Iconisemion) ottogartneri | A. (Icon.) [Icon.ogo] ottogartneri | valid sp. | # subsp. ogoense |
| Aphyosemion (Chromaphyosemion) pamaense | A. (Chrom.) pamaense | valid sp. | |
| Aphyosemion pappenheimi | A. (Chrom.) pappenheimi | = loennbergii | # valid sp. |
| Aphyosemion (Scheelsemion) pascheni pascheni | A. (Sch.) [Sch.pas] pascheni | nomin. subsp. pascheni pascheni | # valid sp. |
| Aphyosemion (Mesoaphyosemion) passaroi | A. (Mes.) [Mes.col] passaroi | valid sp. | |
| Aphyosemion (Aphyosemion) plagitaenium | A. (A.) plagitaenium | valid sp. | |
| Aphyosemion (Chromaphyosemion) poliaki | A. (Chrom.) poliaki | valid sp. | # = volcanum |
| Aphyosemion (Aphyosemion) polli | A. (A.) polli | valid sp. | # = christyi |
| Aphyosemion polychromum | A. (Sch.) [Sch.cal] polychromum | = australe | |
| Aphyosemion preussi | A. (Kath.) preussi | = exiguum | |
| Aphyosemion (Iconisemion) primigenium | A. (Icon.) [Icon.str] primigenium | valid sp. | |
| Aphyosemion (Aphyosemion) pseudoelegans | A. (A.) pseudoelegans | valid sp. | # misidentification |
| Aphyosemion (Mesoaphyosemion) punctatum | A. (Mes.) [Mes.wil] punctatum | valid sp. | |
| Aphyosemion (Chromaphyosemion) punctulatum | A. (Chrom.) punctulatum | valid sp. | |
| Aphyosemion (Iconisemion) pyrophore | A. (Icon.) [Icon.ogo] pyrophore | valid sp. | # subsp. ogoense |
| Aphyosemion (Mesoaphyosemion) raddai | A. (Mes.) [Mes.cam] raddai | valid sp. | |
| Aphyosemion (Aphyosemion) rectogoense | A. (A.) rectogoense | valid sp. | |
| Aphyosemion (Chromaphyosemion) riggenbachi | A. (Chrom.) riggenbachi | valid sp. | |
| Aphyosemion rubrifascium | A. (Kath.) rubrifascium | = bualanum | # = elberti |
| Aphyosemion rubrostictum | A. (Chrom.) rubrostictum | = bitaeniatum | # = bivittatum |
| Aphyosemion (Aphyosemion) schioetzi | A. (A.) schioetzi | valid sp. | # subsp. schioetzi schioetzi |
| Aphyosemion (Iconisemion) schluppi | A. (Icon.) [Icon.thy] schluppi | valid sp. | |
| Aphyosemion (Aphyosemion) schoutedeni | A. (A.) schoutedeni | valid sp. | # = decorsei |
| Aphyosemion schreineri | A. (Rad.) schreineri | = batesii | |
| Aphyosemion (Diapteron) seegersi | A. (D.) seegersi | valid sp. | |
| Aphyosemion simulans | A. (Icon.) [Icon.str] simulans | = escherichi | # = microphtalmum |
| Aphyosemion (Raddaella) splendidum | A. (Rad.) splendidum | valid sp. | # = batesii |
| Aphyosemion (Chromaphyosemion) splendopleure | A. (Chrom.) splendopleure | valid sp. | # = bitaeniatum |
| Aphyosemion (Iconisemion) striatum | A. (Icon.) [Icon.str] striatum | valid sp. | |
| Aphyosemion tessmanni | A. (Kath.) tessmanni | = bualanum | # = elberti |
| Aphyosemion (Aphyosemion) teugelsi | A. (A.) teugelsi | valid sp. | |
| Aphyosemion (Iconisemion) thysi | A. (Icon.) [Icon.thy] thysi | valid sp. | |
| Aphyosemion (Scheelsemion) tirbaki | A. (Sch.) [Sch.her] tirbaki | valid sp. | |
| Aphyosemion unicolor | A. (Sch.) [Sch.cal] unicolor | = calliurum | |
| Aphyosemion unistrigatum | A. (Chrom.) unistrigatum | = loennbergii | |
| Aphyosemion vexillifer | A. (Sch.) [Sch.cal] vexillifer | = calliurum | |
| Aphyosemion (Chromaphyosemion) volcanum | A. (Chrom.) volcanum | valid sp. | # = splendopleure |
| Aphyosemion (Iconisemion) wachtersi | A. (Icon.) [Icon.wac] wachtersi | valid sp. | |
| Aphyosemion (Mesoaphyosemion) wildekampi | A. (Mes.) [Mes.wil] wildekampi | valid sp. | |
| Aphyosemion (Scheelsemion) celiae winifredae | A. (Sch.) [Sch.cal] winifredae | subsp. celiae | # = celiae |
| Aphyosemion (Mesoaphyosemion) hanneloreae wuendschi | A. (Mes.) [Mes.han] wuendschi | subsp. hanneloreae | # = hanneloreae |
| Aphyosemion (Iconisemion) zygaima | A. (Icon.) [Icon.ogo] zygaima | valid sp. |
Why is it important to have dedicated one subgenus to Joergen Scheel ?
Although the dedication of one of the new subgenera, Scheelsemion, is independent to the introductive statement on lumping and splitting, it is tempting to add some after thoughts about it :
- Scheel, whom I have known very well, was a declared lumper (he described very few species and, alone, no name at the genus level) but still he had revolutionized our initial knowledge on Aphyosemion (and on most tropical killifish, hence the herein first dedication of a subgenus to him in killifish as a due token of appreciation) because he was the first to demonstrate with karyological data that speciation was explosive among its components, and at the populational level… but despite that observation he never described alone a cryptic species, i.e., a species that can only be separated from other related components by minor details of color patterns in living male;
- On the contrary, modern researchers do not hesitate a single second to describe a new cryptic species name based on evidenced data that are just a consequence of Scheel's revolution with karyotypes, and that may go to the ultimate state where the difference between the new named cryptic species and the previously described species is so minute that both cannot be separated easily even on photos of the living male (e.g., Scriptaphyosemion wiseae, Simpsonichthys coamazonicus, etc., etc., those 2 examples being emblematic because each corresponds to a description of live fish nearly identical in all details to another species, but with "strong" -at least based on present techniques, such as molecular sequences- scientific evidence… again, no critics over those descriptions and describers, just facts).
Is this situation good or bad ? How would have reacted the pionneer Joergen Scheel with this situation ? Are these new names really useful ? Are those modern researchers with modern techniques just applying Scheel's observations without any risk ? Will there be more and more described cryptic species without an end ? Will ultimately this mean that a population in Scheel's sense (that he wisely recommended to aquarists to keep separate from other related populations for breeding) end up with a species name (e.g., recently 5 populations of Scriptaphyosemion geryi were shown molecularly incompatible, not to speak about the multiple mixed populations of the Aphyosemion elegans superspecies in the Congo basin… or similarly of the Rivulus urophthalmus superspecies in the Amazon basin) ? Will this situation end up with an unavoidable destruction of the nomenclature ? (etc., etc.) Obviously I have no answer to those questions, but still let's believe that there is space, like Scheel always believed (and also any "true" researcher), for new research, new understanding to a biological complexity that becomes more and more complex and farther away while we attempt to touch it… with naming or with no naming, that is the question !
What could be done next ?
Collier (2007) has rightly pointed the uneasy positioning of his "orphan" species, namely hera, hofmanni, in relation to callipteron (Episemion). The facts that all these 3 species are distributed in northern Gabon plateau, not far from each other geographically and that the whole area east of their known distribution in northern Du-Chaillu Massif of hills and small mountains is virgin of detailes collections probably explain those uncertainties. The future prospects for the improvement of our knowledge of the systematics of the genus Aphyosemion most probably lie in that region, all the more that it is precisely where speciation may have once started (at least for the basic lineages Chromaphyosemion and Scheelsemion) and that at its borders there are other atypical fishes without geographical phylogenetic known related counterparts (Plataplochilus terveri, Aphyosemion tirbaki, Aapticheilichthys websteri). Let's hope that entrepreneur collectors, notably passionate hobbyists, will start soon a detailed sampling of the region. Of course if it is not already the case, this is because roads for 4-wheels motors are not available, but there are roads for 2-wheels motors, and bikes with or without motors are fashionable again these days.
The second scope of prospect after this pionneering new study aiming to mimic an available molecular tree while morphological (proportions, meristics) data are untouchable (or parasites) lies probably in applying the same approach to other groups of Killifish : for example, to the genus Fundulus that was well reconfigured recently (we have a diagnosis for the newly named subgenus Wileyichthys, but new rediagnoses are needed for the old subgenera that had been reshuffled like for Aphyosemion), to the genus Aphanius (for which we have molecular data that are inconsistent with morphology), to the genus Epiplatys (in prep. by the present author), to the genus Nothobranchius and its many subgenera (here a complete molecular tree is missing, but rumored as coming soon, and most subgenera have no modern diagnosis), etc. (without forgetting to mention a repeated study of Aphyosemion with new characters notably once a new molecular tree will be obtained with new samples (also in the southern belts in Congo-Zaïre-Angola), and with longer sequences and possibly the whole genome… the cost is decreasing sharply overtime and will become routine, since, for a human being, the whole genome costed 3 years ago US$ 3000 and now it is less than US$ 200 !)
Because this computerized process is accessible to anybody (and notably to aquarists who see their fish alive for long, especially those grouped in specialized associations or study-groups of some genera or even subgenera) but complicated, I shall describe it step by step hereafter (it is free, only a computer is needed and good eyes and time to detail the live fish or color photos… and my eyes are getting old… plus the Philip programme (there are hundreds of such phylogenetic programmes but most are much heavier, more complicated and they use command instructions that are uneasy for no specialists) ; to know more about the programme, go to Philip, written by Joe Felsentein, a nice guy who answers to technical questions by e-mails, a rare asset. Now to process practically :
- go to getme Phylip, select the link to download the Zip Archive (Windows), download it and extract it with a free programme such as IzArc in a dedicated dossier of your computer (the procedure is slightly different with Mac);
- read carefully the instructions and the FAQ page (it is not as easy as a word processor or a spreadsheet processor, but still accessible);
- create and open a new MsExcel file (if you do not own the Excel programme, then download and use the freeware LibreOffice with its included spreadsheet component) and save it anywhere with the name you like (no importance) and select, as a general option of the software, the reference style as L1C1 (not A,B,C, 1,2,3);
- in the file, leave the first 5 lines empty (temporarily);
- in the 6th line and first column, type the formula '=NBCAR(LC(1))' and copy-paste it down for as many lines as you have for studied groups plus your artificial, or not, ancestor (here it was a Fundulopanchax species, for the comparison to all Aphyosemion groups);
- in the 7th line and second column, type the name of your ancestor followed by '$' (here it was AncestorFp$) so that the total number of characters in the first column exactly reads 11 (not less, not more);
- then type down in the second column from line 8 your individual groups at each line (and control that for each the total number of characters in the first column of the corresponding line reads exactly 11 (it is a must that each reads 11 because in the Philip processing of the final matrix it will fail if group names are not exactly sized at 10 characters) ; note : if the number of characters of your group is less than 10, then you may add characters, such as the underscore (e.g., here it was Diapteron_$);
- in line 1 to 6, define each column of characters from column 3 onwards with the definition and possible states of your character (e.g., Max.Size, in line 1), the various states of each character (e.g., 0=very large, over 12 cm, in line 2, 1=large, between 9 and 11 cm, in line 3, 2=medium, between 4.5 cm and 7.5 cm, in line 4, 3=small, under 4 cm, rarely 4.5 cm, in line 5, used for the first character of the Aphyosemion matrix, and eventually in line 6 another 4th character state, not used here);
- then, for each of the character studied, you have to type 0, or 1, or 2, or 3, or in rare cases 4, for each line of each group depending on your observations for each group (that is your added value as a researcher, it is a long meticulous work and a serious work, it takes several dozens of hours, do not improvize because to change things will take even more time and induce errors);
- when you have finished, replace the formula '=NBCAR(LC(1))' from line 7 to the end by a ranked list, 1, 2, 3, etc., starting from the 'ancestor' line;
- your matrix is now finished (but not formatted) with 'n' useful lines from line 7 (not before) to the end and 'nn' items from column 2 (not column 1) with your group names onward to include all your columns filled with characters data;
- next, your matrix must be adapted to a txt file and you will use a freeware named Notepad++ for example (there are hundreds of them, but do NOT use Windows Notepad) or alternatively MSWord (if you do not own the Word programme, then download and use the freeware LibreOffice with its included text processor component);
- copy your matrix (n by nn) into Notepad++, then replace the paragraph (tabulation) sign (which looks like a big blank space character) by nothing, then replace the character '$' by a single standard blank space character and you save the file as _zz1 (_ is to differentiate it from other files, zz is the short name of your work, and 1 is the version number of your matrix) in the subdossier 'exe' of the dossier phylip3.65 in the Philip programme and obtain your final matrix to be processed by Phylip;
- with Windows explorer, go to subdossier 'exe' of the dossier phylip3.65 and double-click on the file 'pars.exe' to open and process Philip with the parsimony analysis;
- with the first line of command that is shown, type '_zz1' (your txt file including your matrix), then 'F', then outfilex (where x is a number for outfile to differentiate it from other further processings);
- then modify settings by selecting letters on various lines (e.g., letter 'O' to select the option 'no, use as outgroup species 1') in an iterative procedure (you have read the instructions before !), then type 'Y' to accept the options and run the programme;
- type F, then outtreex (with 2 't' letters, where x is a number for outtree to differentiate it from other further processings), the window runs and then vanishes;
- with Windows explorer, go again to subdossier 'exe' of the dossier phylip3.65 and double-click on the file named outfilex (x is to be replaced by your chosen number);
- if the first line reads with a number of trees that accounts in many dozens or more, then your work is very heterogeneous (premature) and you'd better go back to your Excel file and check your character states and possibly change or delete some characters definitions or add new characters (and process again);
- if the first line reads with a number of trees that accounts in a few dozens or less, maybe 1 tree or 2 trees, or less than 8 or 24, then your work is not heterogenous (premature), you can note the number of steps of your tree (for information) and you may have a look to the produced tree(s) one after the other (and compare it or them to the molecular tree that you are to mimic or to other previous trees by other researchers with similar groups included);
- then starts a trial and error iterative procedure from Excel to Philip and back again to produce the best tree or the minimum number of trees (by possibly changing or deleting some characters definitions or adding new characters) while using various stages of matrix and files (x being changed accordingly), until you reach your goal;
- that would be the end of your work if the reviewers think it cannot be considered as a new phylogenetic appraisal of the concerned groups because you have been mimicing another work and another (molecular) tree, but not if you are curious or if you obtain more than 1 tree for the parsimony analysis or if your work is designed to propose a new phylogeny, no matter of previous works and trees;
- hence you will have to go further into the analysis of Philip production by using the files named 'consense.exe' (e.g. with Majority rule extended) to build a single concensus tree and 'seqboot.exe', to build the concensus tree with boostrap values (use letter D for 'discrete morphology', then a random number seed, etc.), but that is another story and you will have to explore yourself those new paths of development (the procedure is rather similar to 'pars.exe') and eventually improve your matrix again (notably if you go for the bootstrap strategy and the bootstrap values are very low nearly everywhere);
- when the version of your single tree is finalized (directly or through the concensus process), then you need to copy paste that tree in your MSWord file where your manuscript is being written… good luck !
As a final comment let's hope (idealistically) that one day in the future people, aquarists and researchers, work together in order to define smartly those characters and their states and derive diagnoses for all genera of Killifish !
In total a very important and sensitive newsletter !
Hopefully a boost to our community and a spur to speed up knowledge progress on our (beloved) fishes !
Take care and enjoy the scientific or aquaristic complexity of killifish !
Do not hesitate to ask questions for future Newsletters.
Visit frequently the website www.killi-data.org !
Thank you for your support over the years.
With my kindest regards.
Jean
Literature cited: [the books Killi-Data Series can be ordered here and, now, are even freely sent to K-D-I members as PDF documents]
Huber, J.H. 2012. Reappraisal of the Phylogeny of Rivulus and its Allied focused on External Characters. Killi-Data Series 2012, 9-25, 3 figs., 2 tabs.
Huber, J.H. 2013. Reappraisal of the Phylogeny of the African genus Aphyosemion (Cyprinodontiformes) focused on External Characters, in line with Molecular Data, with new and redefined Subgenera. Killi-Data Series 2013, 4-20, 2 figs.
I am interested in reading other Newsletters, click INFOWEB.


